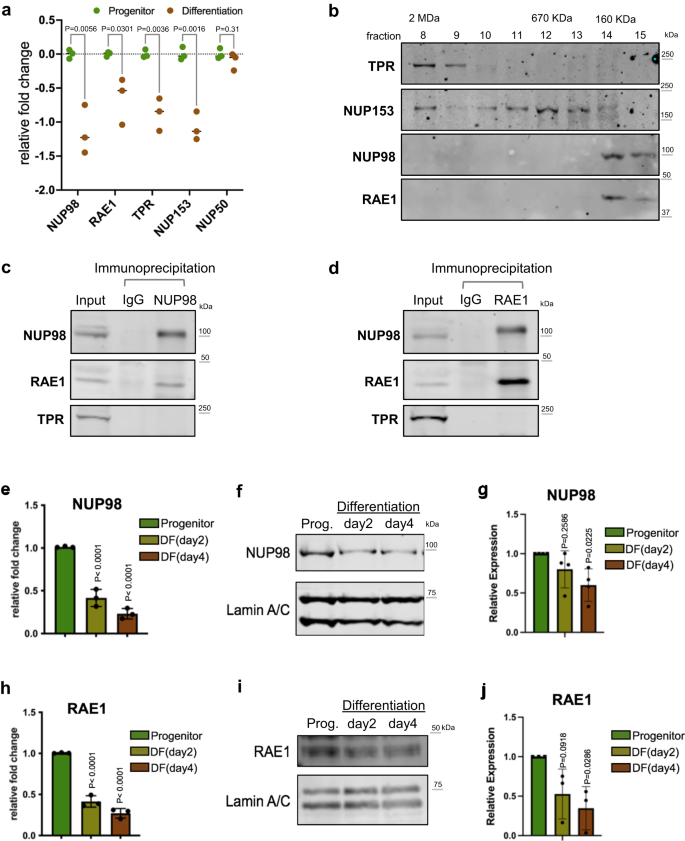
NUP98 and RAE1 are enriched in progenitors and constitute a distinct complex
Between the progenitor-state versus the differentiation state keratinocytes, we identified that 4 out of 5 nuclear-basket NUPs are significantly downregulated in differentiation (Fig. 1a), leveraging the RNA-seq data that we recently generated20. To determine how the enrichment of these nuclear-basket NUPs may influence progenitor maintenance, we asked if these NUPs exist in other complexes inside the nucleus. We extracted the soluble fractions from the nuclei of progenitor-state keratinocytes and performed size-exclusion chromatography. While TPR and NUP153 eluted in a range of earlier fractions, corresponding to larger protein-complex sizes up to 2MDa, NUP98 and RAE1 only eluted in the later fractions around the 160 KDa marker (Fig. 1b). The co-elution of NUP98 and RAE1, in the same fractions corresponding to small protein complexes, suggests that these two proteins may associate with each other independent of each other NUPs. To test this, we performed co-immunoprecipitation using nuclear extraction from progenitor-state keratinocytes. The NUP98 antibody co-immunoprecipitated both NUP98 and RAE1; the RAE1 antibody also co-immunoprecipitated both RAE1 and NUP98. Both co-immunoprecipitations did not enrich other nucleoporins such as TPR (Fig. 1c, d). Since RNA could mediate NUP98’s interactions with other proteins23, we investigated this by comparing NUP98-RAE1 co-immunoprecipitations with or without RNase treatment. Interestingly, the association between NUP98 and RAE1 was minimally affected by RNase (Supplementary Fig. 1a), suggesting that the NUP98-RAE1 interaction does not require an RNA component in this context. In addition, we probed the immunoprecipitated proteins by the NUP98 or RAE1 antibody using mAb414, which recognizes several FG-domain-containing NUPs. Although mAb414 detected multiple bands in the input lysate, this banding pattern was not observed in the immunoprecipitation by the NUP98 or RAE1 antibody (Supplementary Fig. 1b). These data suggest that NUP98 and RAE1 may play a role in the progenitor state independent of other NUPs.
a Relative mRNA expression of nuclear-basket NUPs, comparing the progenitor-state versus the differentiated (day 4) primary human keratinocytes, based on RNA-seq data (multiple unpaired t test, N = 3 biological replicates). b Western blots showing the distribution of nuclear-basket NUPs in the fractions from size-exclusion chromatography (SEC), using the soluble extraction from the nuclei of progenitor-state keratinocytes. The fractions corresponding to the protein standards for SEC are labeled on the top. c, d Western blots showing the co-immunoprecipitation between NUP98 and RAE1 in the soluble extraction from the nuclei (progenitor-state keratinocytes). NUP98 and RAE1 co-immunoprecipitated each other, but not other nuclear-pore subunits such as TPR. e RT-qPCR comparing the relative NUP98 expression at the mRNA level in the progenitor-state, early- (day2) and mid- (day4) differentiation state of keratinocytes (one-way ANOVA with post-hoc test, N = 3 biological replicates, data are represented as mean ± standard deviation). f, g Western blots and quantifications comparing NUP98 protein expression in keratinocyte differentiation, with Lamin A/C used as the loading control (One-way ANOVA with post-hoc test, N = 4, data are represented as mean ± standard deviation). h RT-qPCR comparing the relative RAE1 mRNA expression in keratinocyte differentiation (one-way ANOVA with post-hoc test, data are represented as mean ± standard deviation). i, j Western blots and quantifications comparing RAE1 protein expression levels in keratinocyte differentiation, with Lamin A/C used as the loading control (one-way ANOVA with post-hoc test, N = 3, quantification data are represented as mean ± standard deviation).
To better understand the temporal expression of NUP98 and RAE1, transitioning from the progenitor state towards terminal differentiation, we performed both qRT-PCR and western blotting to quantify their mRNA and protein levels in the time course of calcium-induced keratinocyte differentiation. Significant reduction of NUP98 and RAE1 was detected on Day 2 (early differentiation), and this reduction extended to Day 4 (mid differentiation) of the differentiation time course (Fig. 1e–j). These data indicate that the downregulation of NUP98 and RAE1 is an early event in keratinocyte differentiation, which may play a role in regulating the switch from the progenitor state towards differentiation.
NUP98 or RAE1 knockdown impairs progenitors’ regenerative capacity
To determine if the downregulation of NUP98 or RAE1 promotes the switch from the progenitor state toward differentiation, we leveraged shRNA-mediated knockdown in the progenitor-state keratinocytes, tuning their expression down to a level comparable to the differentiation state. Three independent shRNAs for NUP98 or RAE1 were validated at the mRNA and protein levels (Fig. 2a, b, Supplementary Fig. 2a–d). Reduction of either NUP98 or RAE1 by these 6 shRNAs individually was sufficient to diminish keratinocyte clonogenicity (Fig. 2c–f). To evaluate the roles of NUP98 or RAE1 in influencing progenitors’ regenerative capacity, we performed progenitor competition in skin epidermal regeneration. A 50:50 mix of GFP- or DsRed- expressing progenitor-state keratinocytes were seeded onto devitalized human dermis, raised in liquid-air interface. This organotypic regeneration process completes in a week, forming architecturally faithful human skin epidermis18,19. The GFP-expressing keratinocytes co-expressed control non-targeting shRNA; the DsRed-expressing keratinocytes co-expressed one of the three shRNAs: non-targeting control shRNA, NUP98-targeting shRNA, or RAE1-targeting shRNA. For the epidermis regenerated using keratinocytes both expressing the non-targeting control shRNAs, the red and green fluorescent keratinocytes showed comparable representation in the tissue, indicating that the expression of GFP or DsRed did not differentially influence the progenitors’ regenerative capacity. In contrast, the red keratinocytes co-expressing NUP98 or RAE1 shRNA were outcompeted by the green keratinocytes co-expressing the control shRNA, with diminished representation in the basal progenitor compartment (Fig. 2g, h). These findings suggest that the high expression level of both NUP98 and RAE1 is essential for progenitor maintenance.
a, b RT-qPCR showing knockdown efficiency of 3 independent shRNA targeting NUP98 or RAE1 (one-way ANOVA with post-hoc test, N = 3, data are represented as mean ± standard deviation). c–f Representative images and quantification of clonogenicity assay comparing keratinocytes with NUP98 or RAE1 knockdown versus control (one-way ANOVA with post-hoc test, N = 3, quantification data are represented as mean ± standard deviation.) g Representative images of competition assay in epidermal tissue regeneration. An equal number of keratinocytes expressing DsRed or GFP were mixed seeded onto human dermis. GFP-expressing keratinocytes co-express control shRNA, and DsRed-expressing keratinocytes co-express control shRNA or shRNA targeting NUP98 or RAE1 (scale bar = 125 μm). h Quantification of DsRed or GFP-expressing keratinocytes in the basal layer of the regenerated epidermis, data are represented as mean ± standard deviation. i Venn diagram showing the overlap of NUP98 and RAE1 knockdown differentially expressed genes (Fisher’s exact test, p = 1 × 10−321). j Heatmap showing the relative expression of shared genes with NUP98 or RAE1 knockdown. k Top three gene ontology (GO) terms for the upregulated or downregulated genes shared by NUP98 and RAE1 knockdown, identified by RNA-seq analyses.
To identify the key cellular processes influenced by NUP98 or RAE1 knockdown, we performed transcriptome profiling using RNA-seq. In total, we identified 1493 significantly changed genes with NUP98 knockdown, and 1401 significant changed genes with RAE1 knockdown (p < 0.05, two tailed, Wald test, average fold change for three independent shRNAs >2, and individual shRNA fold change >1.5, Supplementary Data 1, Supplementary Data 2). These two sets share 597 genes (p = 1 × 10−321, Fishers’ exact test), which are altered in the same direction (upregulated or downregulated) with either NUP98 or RAE1 knockdown (Fig. 2i, j). Top gene ontology (GO) terms of the shared upregulated genes include epidermal development and keratinocyte differentiation; Top GO terms of the shared downregulated genes are related to cell division (Fig. 2k). We further identified that 77% of these shared genes are also significantly altered in calcium-induced differentiation (Supplementary Fig. S2e). In addition, we investigated if NUP98 or RAE1 knockdown was sufficient to trigger apoptosis, leveraging two different dyes that are sensitive for mitochondria potential. While the staining was abolished in the positive-control keratinocytes treated with H2O2, keratinocytes with NUP98 or RAE1 knockdown retained the staining similar to the control knockdown (Supplementary Fig. 2f, g), suggesting that the knockdown strategy did not trigger apoptosis. Thus, these findings suggest that the enrichment of NUP98 and RAE1 in the progenitor state is essential for sustaining proliferation and repressing differentiation.
NUP98 binds near the TSSs of key regulators in progenitor maintenance
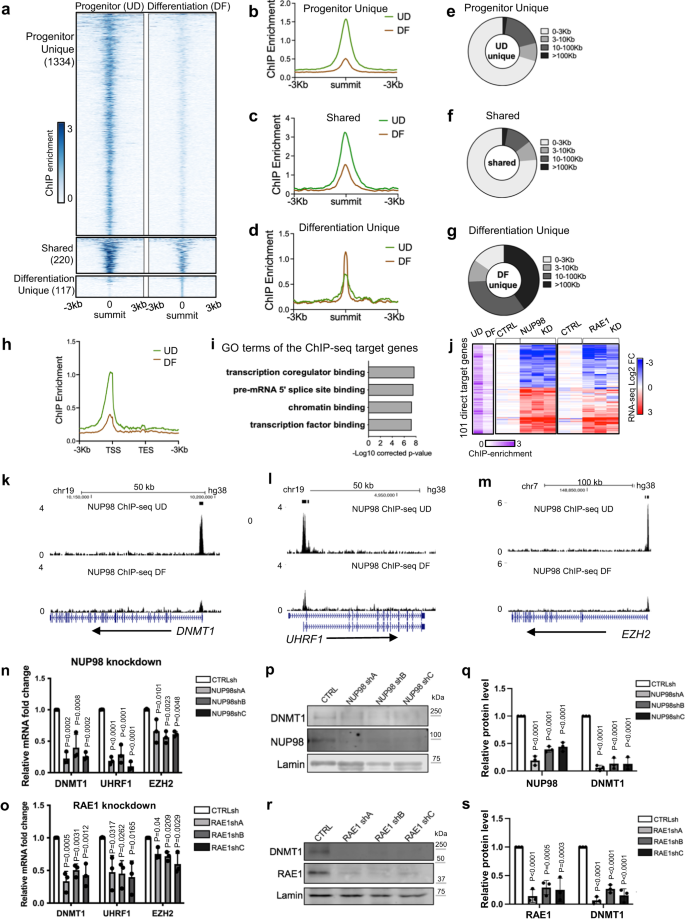
NUP98 has been reported to bind to different genomic regions in the context of different cell types14,24. To investigate how NUP98 genomic binding could influence gene expression in epidermal progenitor maintenance, we performed NUP98 ChIP-seq in keratinocytes. In the progenitor-state keratinocytes, we identified a total of 1554 NUP98 ChIP peaks. In the differentiation state, however, NUP98 binding is reduced across all these regions. The majority (86%, 1334 peaks) of these NUP98 ChIP-seq peaks are unique for the progenitor state, but not for the differentiation state. Only 14% of these peaks (220 peaks) were also called in the differentiation state, yet the ChIP enrichment at these regions is also reduced in differentiation. A small number (117) NUP98 ChIP peaks were identified as unique to the differentiation state, most of which (74.3%) are located at least 10 kb away from the TSSs. In contrast, most peaks (71%) identified in the progenitor state are located within 3 kb from the transcription start sites (Fig. 3a–g, Supplementary Data 3). Thus, the switch from the progenitor state towards differentiation involved an overall reduction of NUP98 genomic binding especially near the transcription start sites (Fig. 3h).
a–d Summit-centered heatmaps and average diagrams comparing NUP98 ChIP enrichment between the progenitor state (UD) and the differentiation state (DF). e–g Pie charts showing the distribution of the NUP98 ChIP-seq peaks, based on their distances to the nearest Transcriptional Start Sites (TSSs). h Average diagram comparing NUP98 ChIP-seq enrichment near the transcription start and end sites of the target genes. i Top Gene Ontology (GO) terms of the nearest genes associated with NUP98 ChIP-seq peaks. j Heatmap showing the 101 genes, featuring NUP98 ChIP-seq enrichment and are significantly changed with NUP98 knockdown. Relative NUP98 ChIP enrichment in UD and DF associated with these gene, and their relative expression with NUP98 or RAE1 knockdown, are included for each of these genes side by side. k–m Browser track examples of NUP98 ChIP-seq enrichment, comparing UD vs. DF. n, o qRT-PCR validation of representative target genes with NUP98 or RAE1 knockdown (one-way ANOVA with post-hoc test, N = 3 biological replicates, data are represented as mean ± standard deviation). p–s Western blot and quantification comparing DNMT1 protein levels with NUP98 or RAE1 knockdown, with Lamin used as the loading control (one-way ANOVA with post-hoc test, N = 3, quantification data are represented as mean ± standard deviation).
We subsequently annotated these genes, which are associated with NUP98 ChIP-seq binding peaks in the progenitor state. Top GO terms are related to transcription co-regulator binding, chromatin binding, and transcription factor binding (Fig. 3i), suggesting that these genes could be upstream regulators of key biological processes. The intersection of the ChIP-seq targets with the NUP98 RNA-seq data identified a total of 101 Direct Target Genes. The majority of these 101 Direct Target Genes also show similar upregulation or downregulation with RAE1 knockdown (Fig. 3j). Interestingly, the downregulated Direct Target Genes include key regulators governing epidermal progenitor maintenance, including the DNA methyltransferase DNMT1 and its recruiter URHF16, the polycomb group protein EZH23 (Fig. 3k–o), in addition to the DNA replication regulators CDT1 and RRM2. These Direct Target Genes also include upregulated genes such as DUSP10 and JARID2, which are less well characterized in the context of keratinocyte differentiation (Supplementary Data 4).
To determine if RAE1 binds to chromatin together with NUP98, we generated a HA-tagged RAE1 construct and expressed it in keratinocytes, as the commercially available antibodies we screened did not yield high-quality ChIP-seq data. HA-RAE1 co-immunoprecipitated NUP98 in the progenitor-state keratinocytes (Supplementary Fig. 3a), confirming that the HA tag does not interfere with RAE1’s association with NUP98. Using double-crosslinking ChIP-seq, we identified that RAE1 is enriched in 83% of the NUP98 ChIP-seq peak regions. The NUP98-RAE1 overlapping ChIP-seq peaks, but not the NUP98 unique peaks, are predominantly located near the TSSs (Supplementary Fig. 3b–f). Among the 101 direct target genes of NUP98, 96 of these genes are also associated with RAE1 ChIP-seq enrichment, including DNMT1 and EZH2 (Supplementary Fig. 3g–i). These findings suggest that NUP98 and RAE1 bind directly near the TSSs to regulate gene expression.
DNMT1 has been previously characterized as a key regulator for human epidermal progenitor maintenance6. We confirmed the drastic downregulation of DNMT1 in keratinocyte differentiation using western blotting (Supplementary Fig. 4a, b). Leveraging the published transcriptome-profiling data of DNMT1 knockdown in progenitor-state keratinocytes6, we compared the differentially expressed genes upon DNMT1 knockdown with the differentially expressed genes upon NUP98 or RAE1 knockdown. The intersection of these data sets identified a total of 236 shared genes (Supplementary Fig. 4c, d). The top GO terms of the upregulated shared genes are associated with epidermal development and keratinocyte differentiation; the downregulated genes are related to cell division and DNA replication (Supplementary Fig. 4e, f). We further validated using western blotting that the DNMT1 protein levels are drastically reduced with NUP98 or RAE1 knockdown, in keratinocytes cultured in the progenitor condition (Fig. 3p–s). Thus, DNMT1 downregulation at least partially accounts for the differentiation induction and proliferation inhibition observed with NUP98 or RAE1 knockdown. Taken together, these data suggest that the chromatin binding of NUP98 and RAE1 is involved in progenitor maintenance, through directly controlling the expression of key proliferation/differentiation regulators such as DNMT1.
NUP98 co-localizes with HDAC1 on chromatin and cooperates with HDAC in gene regulation
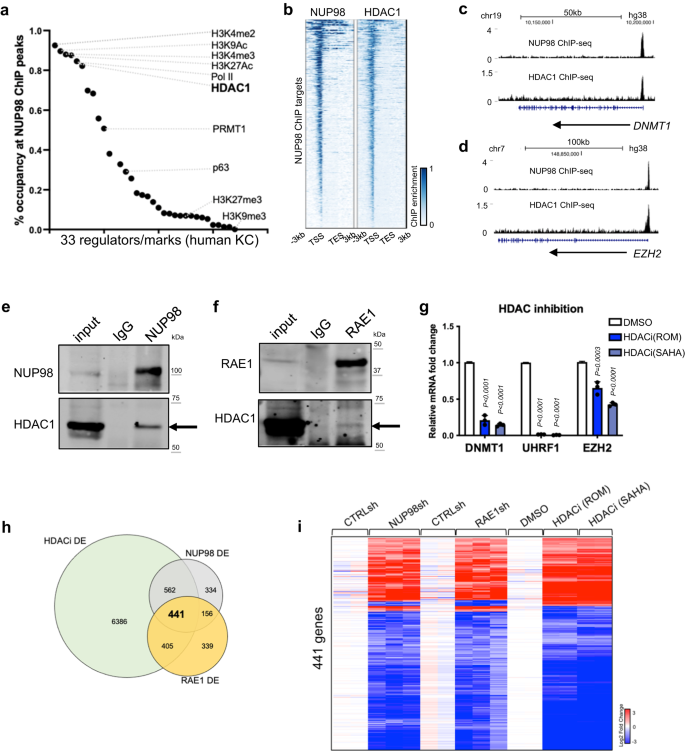
Given NUP98’s chromatin binding to genes encoding key regulators of the proliferation/differentiation process, we investigated potential mechanisms facilitating NUP98’s chromatin binding to these specific genomic regions. Motif search did not uncover specific transcription factors that can explain the majority of the genomic binding sites. We then compared NUP98 ChIP-seq peaks with the ChIP-seq peak files of other transcriptional regulators and histone marks generated using the same cell type of primary human keratinocytes (Fig. 4a, Supplementary Data 5). Consistent with NUP98’s binding near the transcription start sites, NUP98 ChIP-seq peak regions are heavily (>80%) co-occupied by the histone marks (H3K4me2/3, H3K27Ac, H3K9Ac) and Pol II. Interestingly, HDAC1 also co-occupies 82% of all NUP98 ChIP-seq peaks, comparable to Pol II (85%) and much higher than the lineage-specific transcription factor p63 (29%). Similar to NUP98, HDAC1 also binds near the TSSs of the NUP98 target genes, such as DNMT1 and EZH2 (Fig. 4b–d). Thus, HDAC1 stood out as a candidate that could cooperate with NUP98 in chromatin binding and gene regulation.
a Occupancy of other epigenetic marks or regulators in NUP98 ChIP-seq peak regions. b Heatmap comparing NUP98 ChIP-seq and HDAC1 ChIP-seq on NUP98 peaks. c, d Browser tracks showing the co-occupancy of NUP98 and HDAC1 on representative target genes. e, f Crosslinking co-immunoprecipitation using NUP98 or RAE1 antibody to detect HDAC1. g qRT-PCR showing the relative expression of RAE1-NUP98 target genes with HDAC inhibition (one-way ANOVA with post-hoc test, N = 3, data are represented as mean ± standard deviation). hVenn diagram showing the overlap of NUP98, RAE1, and HDACi differentially expressed genes. i Heatmap showing the expression of the differentially expressed genes shared among NUP98 knockdown, RAE1 knockdown, and HDAC inhibition.
This extensive overlap of NUP98 and HDAC1 ChIP-seq peaks suggested that these two proteins could physically associate with each other. We confirmed this using crosslinking immunoprecipitation, that the NUP98 or RAE1 antibody co-immunoprecipitated HDAC1 in keratinocyte lysate (Fig. 4e, f). To determine if HDAC influences gene expression similar to NUP98 and RAE1, we leveraged the HDAC inhibitors Romidepsin (ROM) and SAHA. When added to keratinocytes cultured in the progenitor state, these inhibitors consistently downregulated representative NUP98 direct target genes, such as DNMT1, UHRF1 and EZH2 (Fig. 4g). Using RNA-seq, we further identified that 74% of NUP98-RAE1 target genes are also significantly altered by HDAC inhibition (Fig. 4h, i, Supplementary Data 6). Since p300 was also identified as an interacting protein with NUP98-fusion proteins in hematopoietic malignancies25, we found that p300 inhibition did not drastically alter NUP98’s target gene expression in keratinocytes (Supplementary Fig. 5a), supporting that that HDAC is specifically involved in modulating the target genes controlled by NUP98 and RAE1 in epidermal progenitor maintenance.
Building on the gene expression changes of NUP98 and RAE1’s target genes induced by HDAC inhibition, we further investigated the roles of HDAC1. We designed and validated a total of 3 independent shRNAs targeting HDAC1. Interestingly, all three of these shRNAs consistently downregulated the target genes of NUP98 and RAE1, such as DNMT1 and UHRF1 (Supplementary Fig. 5b). Thus, HDAC1 is involved in regulating the NUP98 and RAE1’s target gene expression in the progenitor state. In the keratinocyte differentiation process, HDAC1’s protein level is slightly reduced, with an average of 54% relative expression on differentiation day 4 as compared to the progenitor state (Supplementary Fig. 5c). Consistently, the HDAC1 ChIP enrichment was only moderately reduced in the differentiation state (Supplementary Fig. 5d). We further compared HDAC1 ChIP-seq enrichment in NUP98 binding sites between the progenitor state versus the differentiation state (day 4, Supplementary Fig. 5e–h). The majority (92.1%) of these sites did not show drastic reduction of HDAC1 binding in differentiation. Only in a very small fraction (7.7%) of these sites, HDAC1 enrichment showed significant reduction (fold change >2, p < 0.05). Thus, HDAC1 ChIP enrichment was only modestly reduced in differentiation in the NUP98 binding sites, in contrast to the drastic reduction of NUP98 in these sites in differentiation, suggesting that NUP98 is not required for maintaining HDAC1’s chromatin binding in their shared binding sites.
NUP98 chromatin binding is dependent on HDAC activity
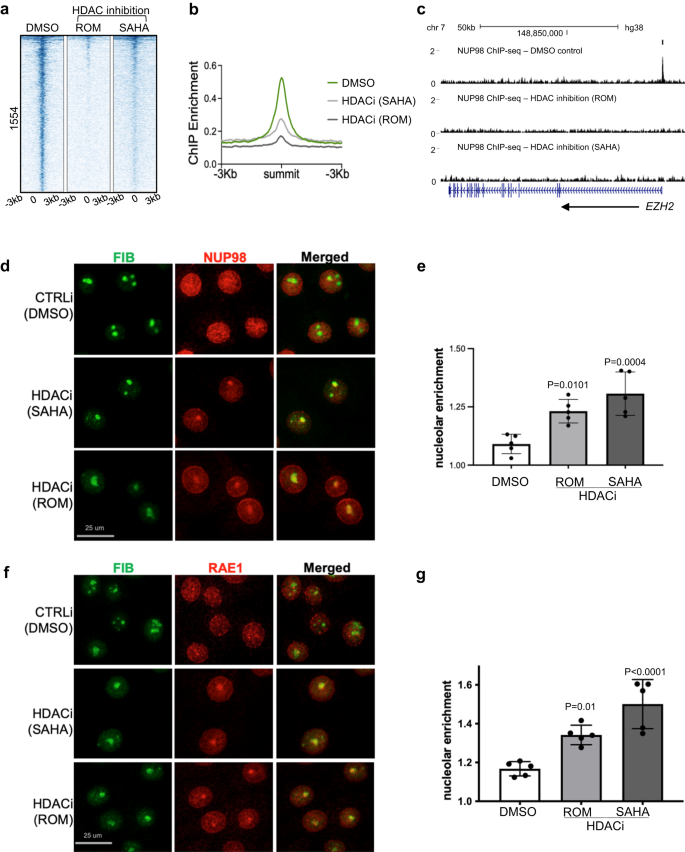
Given the overlap between NUP98 and HDAC1 in genomic binding sites and in gene regulation, we asked if HDAC activity functions upstream to influence NUP98 genomic binding. We performed NUP98 ChIP-seq in keratinocytes with or without HDAC inhibition. Remarkably, the NUP98 ChIP-seq signals were diminished with HDAC inhibition using either SAHA or Romidepsin (Fig. 5a–c), suggesting that NUP98’s genomic binding to its target genes is dependent on HDAC activity.
a Heatmap showing NUP98 ChIP enrichment with HDAC inhibition (using ROM or SAHA) as compared to DMSO control. b Average profile showing NUP98 ChIP enrichment with HDAC inhibition (using ROM or SAHA) as compared to DMSO control. c Browser track showing a representative target gene EZH2, comparing NUP98 ChIP enrichment with HDAC inhibition versus the DMSO control, in keratinocytes cultured in the progenitor state. d Representative images showing keratinocytes co-immunostained with antibodies targeting NUP98 and the nucleolus marker Fibrillarin (FIB), comparing HDAC inhibition versus DMSO control (scale bar = 25 μm). e Quantification of the relative NUP98 enrichment in the nucleolus in HDAC inhibition versus the DMSO control (one-way ANOVA with post-hoc test, N = 5, data are represented as mean ± standard deviation). f, g Co-immunostaining of RAE1 and FIB in keratinocytes treated with HDAC inhibitors versus the DMSO control (scale bar = 25 μm), and quantification (one-way ANOVA with post-hoc test, N = 5, quantification data are represented as mean ± standard deviation).
To determine if NUP98’s subcellular localization was also affected by HDAC inhibition, we performed NUP98 immunofluorescence staining in keratinocytes with HDAC inhibition as compared to DMSO control. While NUP98 showed diffused staining inside the nucleus in the control condition, in addition to its nuclear periphery enrichment, HDAC inhibition by SAHA or Romidespin consistently resulted in 1–2 aggregated foci of NUP98 in each nucleus. Co-staining with the nucleolus marker fibrillin confirmed that the NUP98 localized to the nucleolus (Fig. 5d, e). Similar to NUP98, RAE1 also aggregated to the nucleolus upon HDAC inhibition (Fig. 5f, g).
A recent paper described FUS targeting to the nucleolus under transcriptional stress26. We asked if FUS is involved in NUP98 and RAE1’s nucleolus enrichment. Co-Immunofluorescence staining was performed for both NUP98 and FUS with HDAC inhibition, as compared to the control. While NUP98 was consistently enriched in the nucleolus with HDAC inhibition, no FUS enrichment was detected (Supplementary Fig. 6). These data suggest that NUP98 and RAE1’s nucleolar targeting is independent of the mechanisms involved in targeting FUS to the nucleolus.
NUP98 and RAE1’s nucleolar localization is balanced by HDAC and HAT activities
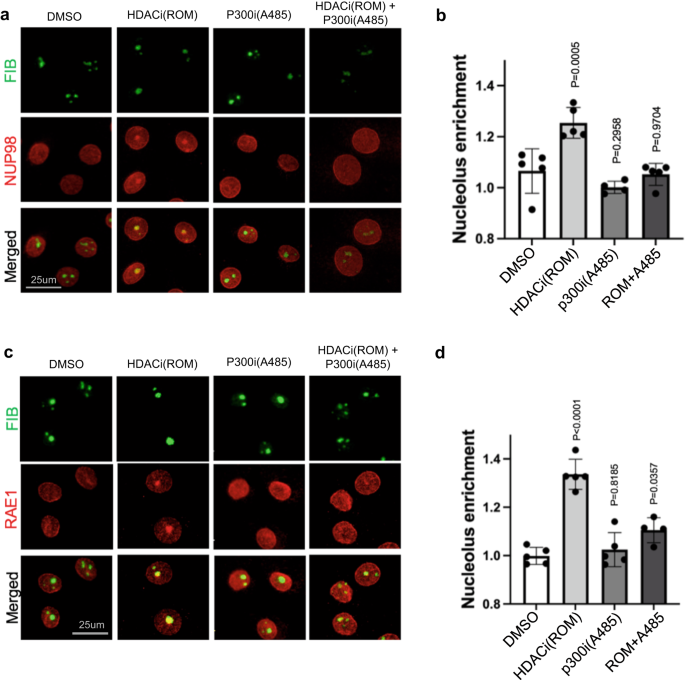
The nucleolar enrichment of NUP98 and RAE1, induced by HDAC inhibition, suggests that protein acetylation could be involved in this process. To test this, we designed an experiment to inhibit protein acetylation in combination with HDAC inhibition, leveraging the p300/CBP HAT inhibitors A485 and C64627,28. As shown in Fig. 6 and Supplementary Fig. 7, HAT inhibition alone did not significantly alter the subnuclear localization of NUP98 or RAE1; however, when the keratinocytes were pre-treated with HAT inhibitors for 6 h, HDAC inhibition was no longer able to induce NUP98 or RAE1’s nucleolar localization. This antagonism between HAT inhibition and HDAC inhibition was consistently observed among the two independent p300/CBP inhibitors (A485 or C646). These data support a role of protein acetylation in mediating the nucleolar localization of NUP98 or RAE1, as a consequence of HDAC inhibition.
a Co-immunostaining of NUP98 and nucleolus marker FIB in keratinocytes treated with HDAC inhibitor, ROM, and/or the HAT inhibitor A485 (scale bar = 25 μm). b quantification of NUP98 nucleolus enrichment in these treatment conditions (one-way ANOVA with post-hoc test, N = 5, data are represented as mean ± standard deviation). c Co-immunostaining of RAE1 and the nucleolus marker FIB in keratinocytes treated with HDAC inhibitor, ROM, and/or the HAT inhibitor A485 (scale bar = 25 μm). d quantification of RAE1 nucleolus enrichment among these conditions (one-way ANOVA with post-hoc test, N = 5, data are represented as mean ± standard deviation).
Interdependence between NUP98 and RAE1 in nucleolar localization upon HDAC inhibition
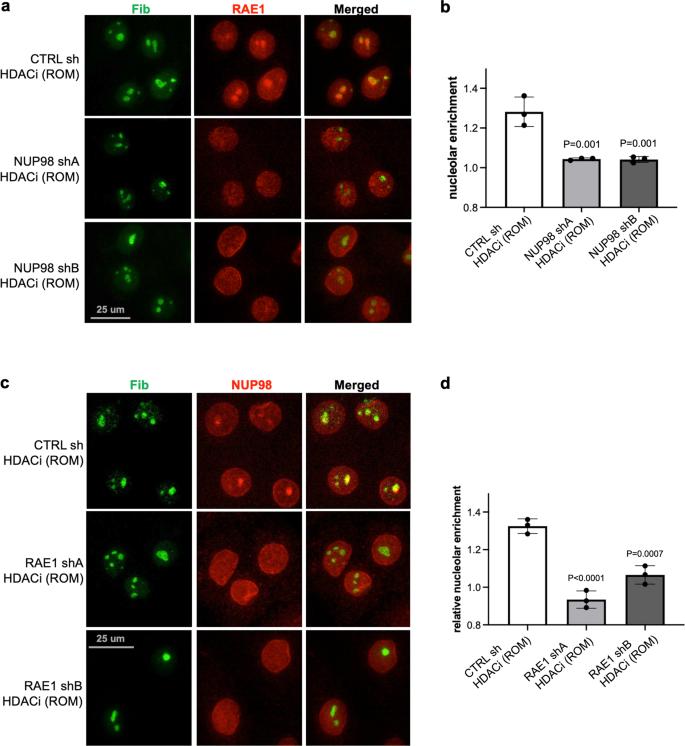
Given the association between NUP98 and RAE1, we further asked if NUP98’s nucleolar targeting upon HDAC inhibition depends on RAE1. While keratinocytes expressing control non-targeting shRNA still showed NUP98 aggregation with HDAC inhibition, this nucleolar aggregation was abolished in keratinocytes expressing shRNAs targeting RAE1 (Fig. 7a, b). Thus, the nucleolar enrichment of NUP98 requires the intact function of RAE1. We subsequently investigated if RAE1’s nucleolar targeting upon HDAC inhibition also depended on NUP98. We treated keratinocytes expressing either NUP98 shA, NUP98 shB or control non-targeting shRNA with the HDAC inhibitor ROM. RAE1’s nucleolar targeting was abolished in keratinocytes expressing NUP98 shRNA, but not the non-targeting control. Using two independent shRNAs targeting NUP98, RAE1 also did not target to the nucleolus (Fig. 7c, d). RAE1 knockdown alone did not induce NUP98 nucleolar localization, and NUP98 knockdown did not induce RAE1 nucleolar localization, without HDAC inhibition (Supplementary Fig. 8). These findings indicate that NUP98 and RAE1 depend on each other to localize to the nucleolus upon HDAC inhibition.
a, b Representative images showing co-immunostaining of RAE1 and the nucleolus marker FIB in keratinocytes treated with NUP98 knockdown versus control knockdown (scale bar = 25 μm), and quantifications of RAE1 nucleolar enrichment among these conditions (one-way ANOVA with post-hoc test, N = 3, quantification data are represented as mean ± standard deviation). c, d Co-immunostaining of NUP98 and the nucleolus marker FIB in keratinocytes treated with RAE1 knockdown versus control knockdown (scale bar = 25 μm), and quantifications of NUP98 nucleolar enrichment among these conditions (one-way ANOVA with post-hoc test, N = 3, quantification data are represented as mean ± standard deviation).
Taken together, these findings suggest a model that NUP98 and RAE1 promote progenitor maintenance by directly binding to and controlling the expression of key epigenetic regulators under two conditions: elevated expression and HDAC activity. The elevated expression in the progenitor state allows the presence of a soluble intranuclear pool of NUP98 and RAE1, allowing chromatin binding in addition to their nuclear-pore association; HDAC activity is also essential for antagonizing the nucleolar targeting for NUP98 and RAE1, promoting NUP98 genomic targeting to the key epigenetic regulators for self-renewal (Fig. 8).
NUP98 and RAE1 support epidermal progenitor maintenance through binding to chromatin near the transcription start sites of key target genes such as the epigenetic repressor DNMT1. NUP98 and RAE1’s roles in progenitor maintenance depend on two factors: high expression and HDAC activity. The high expression of NUP98 and RAE1 provides the chromatin-binding pool, in addition to their nuclear-pore incorporation; HDAC activity supports NUP98’s binding on chromatin near the TSSs of key target genes, antagonizing the nucleolar localization of NUP98 and RAE1.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Automotive / EVs, Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- BlockOffsets. Modernizing Environmental Offset Ownership. Access Here.
- Source: https://www.nature.com/articles/s42003-023-05043-2